2023
Semi-Implicit Denoising Diffusion Models (SIDDMs)
Yanwu Xu, Mingming Gong, Shaoan Xie, Wei Wei, Matthias Grundmann, Kayhan Batmanghelich†, Tingbo Hou†
37th Conference on Neural Information Processing Systems (NeurIPS 2023)
[pdf]
Abstract
Despite the proliferation of generative models, achieving fast sampling during inference without compromising sample diversity and quality remains challenging. Existing models such as Denoising Diffusion Probabilistic Models (DDPM) deliver high-quality, diverse samples but are slowed by an inherently high number of iterative steps. The Denoising Diffusion Generative Adversarial Networks (DDGAN) attempted to circumvent this limitation by integrating a GAN model for larger jumps in the diffusion process. However, DDGAN encountered scalability limitations when applied to large datasets. To address these limitations, we introduce a novel approach that tackles the problem by matching implicit and explicit factors. More specifically, our approach involves utilizing an implicit model to match the marginal distributions of noisy data and the explicit conditional distribution of the forward diffusion. This combination allows us to effectively match the joint denoising distributions. Unlike DDPM but similar to DDGAN, we do not enforce a parametric distribution for the reverse step, enabling us to take large steps during inference. Similar to the DDPM but unlike DDGAN, we take advantage of the exact form of the diffusion process. We demonstrate that our proposed method obtains comparable generative performance to diffusion-based models and vastly superior results to models with a small number of sampling steps.
Abstract
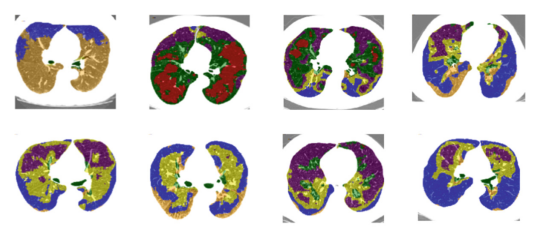
Large-scale volumetric medical images with annotation are rare, costly, and time prohibitive to acquire. Self-supervised learning (SSL) offers a promising pre-training and feature extraction solution for many downstream tasks, as it only uses unlabeled data. Recently, SSL methods based on instance discrimination have gained popularity in the medical imaging domain. However, SSL pre-trained encoders may use many clues in the image to discriminate an instance that are not necessarily disease-related. Moreover, pathological patterns are often subtle and heterogeneous, requiring the ability of the desired method to represent anatomy-specific features that are sensitive to abnormal changes in different body parts. In this work, we present a novel SSL framework, named DrasCLR, for 3D lung CT images to overcome these challenges. We propose two domain-specific contrastive learning strategies: one aims to capture subtle disease patterns inside a local anatomical region, and the other aims to represent severe disease patterns that span larger regions. We formulate the encoder using conditional hyper-parameterized network, in which the parameters are dependant on the anatomical location, to extract anatomically sensitive features. Extensive experiments on large-scale datasets of lung CTscans show that our method improves the performance of many downstream prediction and segmentation tasks. The patient-level representation improves the performance of the patient survival prediction task. We show how our method can detect emphysema subtypes via dense prediction. We demonstrate that fine-tuning the pre-trained model can significantly reduce annotation efforts without sacrificing emphysema detection accuracy. Our ablation study highlights the importance of incorporating anatomical context into the SSL framework.
Abstract
Deep Neural Networks (DNNs) are prone to learning spurious features that correlate with the label during training but are irrelevant to the learning problem. This hurts model generalization and poses problems when deploying them in safety-critical applications. This paper aims to better understand the effects of spurious features through the lens of the learning dynamics of the internal neurons during the training process. We make the following observations: (1) While previous works highlight the harmful effects of spurious features on the generalization ability of DNNs, we emphasize that not all spurious features are harmful. Spurious features can be “benign” or “harmful” depending on whether they are “harder” or “easier” to learn than the core features for a given model. This definition is model and dataset dependent. (2) We build upon this premise and use instance difficulty methods (like Prediction Depth (Baldock et al., 2021)) to quantify “easiness” for a given model and to identify this behavior during the training phase. (3) We empirically show that the harmful spurious features can be detected by observing the learning dynamics of the DNN’s early layers. In other words, easy features learned by the initial layers of a DNN early during the training can (potentially) hurt model generalization. We verify our claims on medical and vision datasets, both simulated and real, and justify the empirical success of our hypothesis by showing the theoretical connections between Prediction Depth and information-theoretic concepts like V-usable information (Ethayarajh et al., 2021). Lastly, our experiments show that monitoring only accuracy during training (as is common in machine learning pipelines) is insufficient to detect spurious features. We, therefore, highlight the need for monitoring early training dynamics using suitable instance difficulty metrics.
Abstract
Studying small effects or subtle neuroanatomical variation requires large-scale sample size data. As a result, combining neuroimaging data from multiple datasets is necessary. Variation in acquisition protocols, magnetic field strength, scanner build, and many other non-biologically related factors can introduce undesirable bias into studies. Hence, harmonization is required to remove the bias-inducing factors from the data. ComBat is one of the most common methods applied to features from structural images. ComBat models the data using a hierarchical Bayesian model and uses the empirical Bayes approach to infer the distribution of the unknown factors. The empirical Bayes harmonization method is computationally efficient and provides valid point estimates. However, it tends to underestimate uncertainty. This paper investigates a new approach, fully Bayesian ComBat, where Monte Carlo sampling is used for statistical inference. When comparing fully Bayesian and empirical Bayesian ComBat, we found Empirical Bayesian ComBat more effectively removed scanner strength information and was much more computationally efficient. Conversely, fully Bayesian ComBat better preserved biological disease and age-related information while performing more accurate harmonization on traveling subjects. The fully Bayesian approach generates a rich posterior distribution, which is useful for generating simulated imaging features for improving classifier performance in a limited data setting. We show the generative capacity of our model for augmenting and improving the detection of patients with Alzheimer’s disease. Posterior distributions for harmonized imaging measures can also be used for brain-wide uncertainty comparison and more principled downstream statistical analysis.
Distilling Blackbox to Interpretable Models for Efficient Transfer Learning
Shantanu Ghosh, Ke Yu, Kayhan Batmanghelich
26th International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI 2023)
[pdf][code][project page]
Abstract
Building generalizable AI models is one of the primary challenges in the healthcare domain. While radiologists rely on generalizable descriptive rules of abnormality, Neural Network (NN) models suffer even with a slight shift in input distribution (e.g., scanner type). Fine-tuning a model to transfer knowledge from one domain to another requires a significant amount of labeled data in the target domain. In this paper, we develop an interpretable model that can be efficiently fine-tuned to an unseen target domain with minimal computational cost. We assume the interpretable component of NN to be approximately domain-invariant. However, interpretable models typically underperform compared to their Blackbox (BB) variants. We start with a BB in the source domain and distill it into a mixture of shallow interpretable models using human-understandable concepts. As each interpretable model covers a subset of data, a mixture of interpretable models achieves comparable perfor- mance as BB. Further, we use the pseudo-labeling technique from semi-supervised learning (SSL) to learn the concept classifier in the target domain, followed by fine-tuning the interpretable models in the target domain. We evaluate our model using a real-life large-scale chest-X-ray (CXR) classification dataset.
Abstract
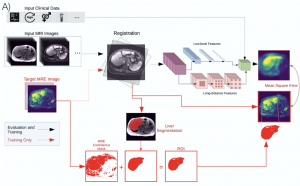
Magnetic resonance elastography (MRE) is a medical imaging modality that non-invasively quantifies tissue stiffness (elasticity) and is commonly used for diagnosing liver fibrosis. Constructing an elasticity map of tissue requires solving an inverse problem involving a partial differential equation (PDE). Current numerical techniques to solve the inverse problem are noise-sensitive and require explicit specification of physical relationships. In this work, we apply physics-informed neural networks to solve the inverse problem of tissue elasticity reconstruction. Our method does not rely on numerical differentiation and can be extended to learn relevant correlations from anatomical images while respecting physical constraints. We evaluate our approach on simulated data and in vivo data from a cohort of patients with non-alcoholic fatty liver disease (NAFLD). Compared to numerical baselines, our method is more robust to noise and more accurate on realistic data, and its performance is further enhanced by incorporating anatomical information.
Deep Learning Integration of Chest CT Imaging and Gene Expression Identifies Novel Aspects of COPD
Junxiang Chen, Zhonghui, Xu, Li Sun, Ke Yu, Craig P. Hersh, Adel Boueiz, John Hokanson, Frank C. Sciurba, Edwin K. Silverman, Peter J. Castaldi, Kayhan Batmanghelich
to appear in Chronic Obstructive Pulmonary Diseases: Journal of the COPD Foundation (medRxiv:doi.org/10.1101/2022.09.26.22280242)
[pdf][code]
Abstract
Rationale Chronic obstructive pulmonary disease (COPD) is characterized by pathologic changes in the airways, lung parenchyma, and persistent inflammation, but the links between lung structural changes and patterns of systemic inflammation have not been fully described.
Objectives To identify novel relationships between lung structural changes measured by chest computed tomography (CT) and systemic inflammation measured by blood RNA sequencing.
Methods CT scan images and blood RNA-seq gene expression from 1,223 subjects in the COPDGene study were jointly analyzed using deep learning to identify shared aspects of inflammation and lung structural changes that we refer to as Image-Expression Axes (IEAs). We related IEAs to COPD-related measurements and prospective health outcomes through regression and Cox proportional hazards models and tested them for biological pathway enrichment.
Measurements and Main Results We identified two distinct IEAs: IEAemph captures an emphysema-predominant process with a strong positive correlation to CT emphysema and a negative correlation to FEV1 and Body Mass Index (BMI); IEAairway captures an airway-predominant process with a positive correlation to BMI and airway wall thickness and a negative correlation to emphysema. Pathway enrichment analysis identified 29 and 13 pathways significantly associated with IEAemph and IEAairway, respectively (adjusted p<0.001).
Conclusions Integration of CT scans and gene expression data identified two IEAs that capture distinct inflammatory processes associated with emphysema and airway-predominant COPD.
Scientific Knowledge on the Subject Chronic obstructive pulmonary disease (COPD) is characterized by lung structural changes and has a prominent systemic inflammatory component, but the links between lung structural changes and patterns of systemic inflammation in COPD have not been fully described.
What This Study Adds to the Field We identified novel relationships between lung structural changes and systemic inflammation by simultaneously analyzing CT scans and blood RNA-sequencing gene expression using deep learning models. We identified two distinct Image-Expression Axes (IEAs) that characterize different inflammatory processes associated with emphysema and airway predominant COPD.
Dividing and Conquering a BlackBox to a Mixture of Interpretable Models: Route, Interpret, Repeat
Shantanu Ghosh, Ke Yu, Forough Arabshahi, Kayhan Batmanghelich
The Fortieth International Conference on Machine Learning (ICML)
[pdf][code][project page]
Abstract
ML model design either starts with an interpretable model or a Blackbox and explains it post hoc. Blackbox models are flexible but difficult to explain, while interpretable models are inherently explainable. Yet, interpretable models require extensive ML knowledge and tend to be less flexible, potentially underperforming than their Blackbox equivalents. This paper aims to blur the distinction between a post hoc explanation of a Blackbox and constructing interpretable models. Beginning with a Blackbox, we iteratively carve out a mixture of interpretable models and a residual network. The interpretable models identify a subset of samples and explain them using First Order Logic (FOL), providing basic reasoning on concepts from the Blackbox. We route the remaining samples through a flexible residual. We repeat the method on the residual network until all the interpretable models explain the desired proportion of data. Our extensive experiments show that our route, interpret, and repeat approach (1) identifies a richer diverse set of instance-specific concepts with high concept completeness via interpretable models by specializing in various subsets of data without compromising in performance, (2) identifies the relatively “harder” samples to explain via residuals, (3) outperforms the interpretable by-design models by significant margins during test-time interventions, (4) can be used to fix the shortcut learned by the original Blackbox.
Abstract
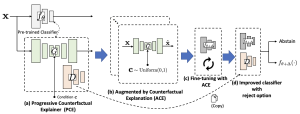
A highly accurate but overconfident model is ill-suited for deployment in critical applications such as healthcare and autonomous driving. The classification outcome should reflect a high uncertainty on ambiguous in-distribution samples that lie close to the decision boundary. The model should also refrain from making overconfident decisions on samples that lie far outside its training distribution, far-out-of-distribution (far-OOD), or on unseen samples from novel classes that lie near its training distribution (near-OOD). This paper proposes an application of counterfactual explanations in fixing an over-confident classifier. Specifically, we propose to fine-tune a given pre-trained classifier using augmentations from a counterfactual explainer (ACE) to fix its uncertainty characteristics while retaining its predictive performance. We perform extensive experiments with detecting far-OOD, near-OOD, and ambiguous samples. Our empirical results show that the revised model have improved uncertainty measures, and its performance is competitive to the state-of-the-art methods.
Explaining the Black-box Smoothly – A Counterfactual Approach
Sumedha Singla, Motahhare Eslami, Brian Pollack, Stephen Wallace, Kayhan Batmanghelich
Medical Image Analysis, vol 84, Feb 23 (Preprint:arXiv:2101.04230)
[pdf][tbd]
Abstract
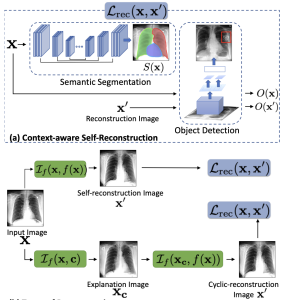
We propose a BlackBox Counterfactual Explainer, designed to explain image classification models for medical applications. Classical approaches (e.g., saliency maps) that assess feature importance do not explain how imaging features in important anatomical regions are relevant to the classification decision. Such reasoning is crucial for transparent decision-making in healthcare applications. Our framework explains the decision for a target class by gradually exaggerating the semantic effect of the class in a query image. We adopted a Generative Adversarial Network (GAN) to generate a progressive set of perturbations to a query image, such that the classification decision changes from its original class to its negation. Our proposed loss function preserves essential details (e.g., support devices) in the generated images. We used counterfactual explanations from our framework to audit a classifier trained on a chest x-ray dataset with multiple labels. Clinical evaluation of model explanations is a challenging task. We proposed clinically-relevant quantitative metrics such as cardiothoracic ratio and the score of a healthy costophrenic recess to evaluate our explanations. We used these metrics to quantify the counterfactual changes between the populations with negative and positive decisions for a diagnosis by the given classifier. We conducted a human-grounded experiment with diagnostic radiology residents to compare different styles of explanations (no explanation, saliency map, cycleGAN explanation, and our counterfactual explanation) by evaluating different aspects of explanations: (1) understandability, (2) classifier’s decision justification, (3) visual quality, (d) identity preservation, and (5) overall helpfulness of an explanation to the users. Our results show that our counterfactual explanation was the only explanation method that significantly improved the users’ understanding of the classifier’s decision compared to the no-explanation baseline. Our metrics established a benchmark for evaluating model explanation methods in medical images. Our explanations revealed that the classifier relied on clinically relevant radiographic features for its diagnostic decisions, thus making its decision-making process more transparent to the end-user.
2022
Automated Detection of Premalignant Oral Lesions on Whole Slide Images Using Convolutional Neural Networks
Yingci Liu, Elizabeth Bilodeau, Brian Pollack, Kayhan Batmanghelich
Oral Oncology
[pdf][tbd]
Abstract
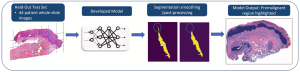
Introduction: Oral epithelial dysplasia (OED) is a precursor lesion to oral squamous cell carcinoma, a disease with a reported overall survival rate of 56 percent across all stages. Accurate detection of OED is critical as progression to oral cancer can be impeded with complete excision of premalignant lesions. However, previous research has demonstrated that the task of grading of OED, even when performed by highly trained experts, is subject to high rates of reader variability and misdiagnosis. Thus, our study aims to develop a convolutional neural network (CNN) model that can identify regions suspicious for OED whole-slide pathology images.
Methods: During model development, we optimized key training hyperparameters, including loss function on 112 pathologist annotated cases between the training and validation sets. Then, we compared OED segmentation and classification metrics between two well-established CNN architectures for medical imaging, DeepLabv3+ and UNet++. To further assess generalizability, we assessed case-level performance of a held-out test set of 44 whole-slide images.
Results: DeepLabv3+ outperformed UNet++ in overall accuracy, precision, and segmentation metrics in a 4-fold cross validation study. When applied to the held-out test set, our best performing DeepLabv3+ model achieved an overall accuracy and F1-Score of 93.3 percent and 90.9 percent, respectively.
Conclusion: The present study trained and implemented a CNN-based deep learning model for the identification and segmentation of oral epithelial dysplasia (OED) with reasonable success. Computer assisted detection was shown to be feasible in detecting premalignant/precancerous oral lesions, laying the groundwork for eventual clinical implementation.
Abstract
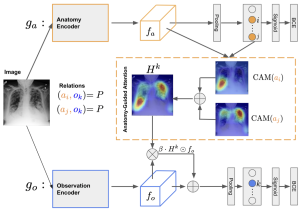
Creating a large-scale dataset of abnormality annotation on medical images is a labor-intensive and costly task. Leveraging weak supervision from readily available data, such as radiology reports can compensate lack of large-scale data for anomaly detection methods. However, most of the current methods only use image-level pathological observations, failing to utilize the relevant anatomy mentions in reports. Furthermore, Natural Language Processing (NLP)-mined weak labels are noisy due to label sparsity and linguistic ambiguity. We propose an Anatomy- Guided chest X-ray Network (AGXNet) to address these issues of weak annotation. Our framework consists of a cascade of two networks, one responsible for identifying anatomical abnormalities and the second responsible for pathological observations. The critical component in our framework is an anatomy-guided attention module that aids the down-stream observation network in focusing on the relevant anatomical regions generated by the anatomy network. We use Positive Unlabeled (PU) learning to account for the fact that lack of mention does not necessarily mean a negative label. Our quantitative and qualitative results on the MIMIC-CXR dataset demonstrate the effectiveness of AGXNet in disease and anatomical abnormality localization. Experiments on the NIH Chest X-ray dataset show that the learned feature representations are transferable and can achieve the state-of-the-art performances in disease classification and competitive disease localization results.
Abstract
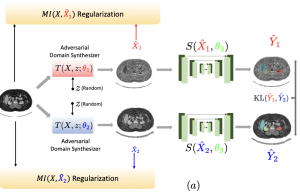
An organ segmentation method that can generalize to un- seen contrasts and scanner settings can significantly reduce the need for retraining of deep learning models. Domain Generalization (DG) aims to achieve this goal. However, most DG methods for segmentation require training data from multiple domains during training. We propose a novel adversarial domain generalization method for organ segmentation trained on data from a single domain. We synthesize the new domains via learning an adversarial domain synthesizer (ADS) and presume that the synthetic domains cover a large enough area of plausible distributions so that unseen domains can be interpolated from synthetic domains. We propose a mutual information regularizer to enforce the semantic consistency between images from the synthetic domains, which can be estimated by patch-level contrastive learning. We evaluate our method for various organ segmentation for unseen modalities, scanning protocols, and scanner sites.
Abstract
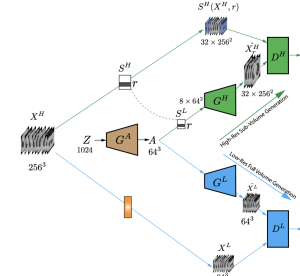
Generative Adversarial Networks (GAN) have many potential medical imaging applications, including data augmentation, domain adaptation, and model explanation. Due to the limited embedded memory of Graphical Processing Units (GPUs), most current 3D GAN models are trained on low-resolution medical images. In this work, we propose a novel end-to-end GAN architecture that can generate high-resolution 3D images. We achieve this goal by separating training and inference. During training, we adopt a hierarchical structure that simultaneously generates a low-resolution version of the image and a randomly selected sub-volume of the high-resolution image. The hierarchical design has two advantages: First, the memory demand for training on high-resolution images is amortized among subvolumes. Furthermore, anchoring the high-resolution subvolumes to a single low-resolution image ensures anatomical consistency between subvolumes. During inference, our model can directly generate full high-resolution images. We also incorporate an encoder with a similar hierarchical structure into the model to extract features from the images. Experiments on 3D thorax CT and brain MRI demonstrate that our approach outperforms state of the art in image generation, image reconstruction, and clinical-relevant variables prediction.
Abstract
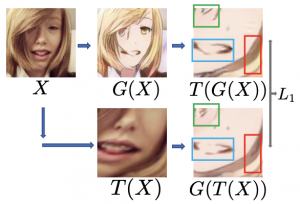
Unpaired image-to-image translation (I2I) is an ill-posed problem, as an infinite number of translation functions can map the source domain distribution to the target distribution. Therefore, much effort has been put into designing suitable constraints, e.g., cycle consistency (CycleGAN), geometry consistency (GCGAN), and contrastive learning-based constraints (CUTGAN), that help better pose the problem. However, these well-known constraints have limitations: (1) they are either too restrictive or too weak for specific I2I tasks; (2) these methods result in content distortion when there is a significant spatial variation between the source and target domains. This paper proposes a universal regularization technique called maximum spatial perturbation consistency (MSPC), which enforces a spatial perturbation function ($T$) and the translation operator ($G$) to be commutative (i.e., $T \circ G = G \circ T $). In addition, we introduce two adversarial training components for learning the spatial perturbation function. The first one lets $T$ compete with $G$ to achieve maximum perturbation. The second one lets $G$ and $T$ compete with discriminators to align the spatial variations caused by the change of object size, object distortion, background interruptions, etc. Our method outperforms the state-of-the-art methods on most I2I benchmarks. We also introduce a new benchmark, namely the front face to profile face dataset, to emphasize the underlying challenges of I2I for real-world applications. We finally perform ablation experiments to study the sensitivity of our method to the severity of spatial perturbation and its effectiveness for distribution alignment.
Knowledge Distillation via Constrained Variational Inference
Ardavan Saeedi, Yuria Utsumi, Li Sun, Kayhan Batmanghelich, Li-wei H. Lehman
Association for the Advancement of Artificial Intelligence (AAAI 2022)
[pdf][tbd]
Abstract
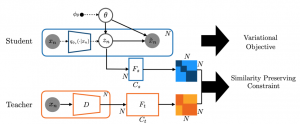
Knowledge distillation has been used to capture the knowledge of a teacher model and distill it into a student model with some desirable characteristics such as being smaller, more efficient, or more generalizable. In this paper, we propose a framework for distilling the knowledge of a powerful discriminative model such as a neural network into commonly used graphical models known to be more interpretable (e.g., topic models, autoregressive Hidden Markov Models). Posterior of latent variables in these graphical models (e.g., topic proportions in topic models) is often used as feature representation for predictive tasks. However, these posterior-derived features are known to have poor predictive performance compared to the features learned via purely discriminative approaches. Our framework constrains variational inference for posterior variables in graphical models with a similarity preserving constraint. This constraint distills the knowledge of the discriminative model into the graphical model by ensuring that input pairs with (dis)similar representation in the teacher model also have (dis)similar representation in the student model. By adding this constraint to the variational inference scheme, we guide the graphical model to be a reasonable density model for the data while having predictive features which are as close as possible to those of a discriminative model. To make our framework applicable to a wide range of graphical models, we build upon the Automatic Differentiation Variational Inference (ADVI), a black-box inference framework for graphical models. We demonstrate the effectiveness of our framework on two real-world tasks of disease subtyping and disease trajectory modeling.
2021
Abstract
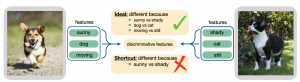
The generalization of representations learned via contrastive learning depends crucially on what features of the data are extracted. However, we observe that the contrastive loss does not always sufficiently guide which features are extracted, a behavior that can negatively impact the performance on downstream tasks via “shortcuts”, i.e., by inadvertently suppressing important predictive features. We find that feature extraction is influenced by the difficulty of the so-called instance discrimination task (i.e., the task of discriminating pairs of similar points from pairs of dissimilar ones). Although harder pairs improve the representation of some features, the improvement comes at the cost of suppressing previously well-represented features. In response, we propose implicit feature modification (IFM), a method for altering positive and negative samples in order to guide contrastive models towards capturing a wider variety of predictive features. Empirically, we observe that IFM reduces feature suppression, and as a result, improves performance on vision and medical imaging tasks.
Deep Learning Prediction of Voxel-Level Liver Stiffness in Patients with Nonalcoholic Fatty Liver Disease
B. L. Pollack*, K. Batmanghelich*, S. S. Cai, E. Gordon, S. Wallace, R. Catania, C. Morillo-Hernandez, A. Furlan, A. A. Borhani
Radiology: Artificial Intelligence (doi: 10.1148/ryai.2021200274)
[pdf][code]
Abstract
To reconstruct virtual MR elastography (MRE) images based on traditional MRI inputs with a machine learning algorithm. In this single-institution retrospective study, 149 patients (mean age, 58 ± 12, 71 men) with nonalcoholic fatty liver disease (NAFLD) who underwent MRI and MRE between January 2016 and January 2019 were evaluated. Nine conventional MRI sequences and clinical data were used to train a convolutional neural network (CNN) to reconstruct MRE images at the per-voxel level. The architecture was further modified to accept multichannel three-dimensional inputs and to allow inclusion of clinical and demographic information. Liver stiffness and fibrosis category (F0-F4) of reconstructed images were assessed using voxel-level and patient-level agreement by correlation, sensitivity, and specificity calculations, as well as classification by receiver operator characteristic analyses, and Dice score to evaluate hepatic stiffness locality. The model for predicting liver stiffness incorporated 4 image sequences (precontrast T1-weighted liver acquisition with volume acquisition [LAVA]-water and LAVA-fat, 120-second delay T1-weighted LAVA-water, and T2-weighted) and clinical data. The model had a patient-level and voxel-level correlation of 0.50 ± 0.05 and 0.34 ± 0.03. Using a stiffness threshold of 3.54 kPa to binarize classification into no or mild fibrosis (F0-F1) versus clinically significant fibrosis (F2-F4), the model had a sensitivity of 80% ± 4%, specificity of 75% ± 5%, accuracy of 78% ± 3%, area under the curve of 84 ± 0.04, and a Dice score of 0.74. The generation of virtual elastography images is feasible using conventional MRI and clinical data with a machine learning algorithm.
Abstract
Vessel segmentation is an essential task in many clinical applications. Although supervised methods have achieved state-of-art performance, acquiring expert annotation is laborious and mostly limited for two-dimensional datasets with small sample size. On the contrary, unsupervised methods rely on handcrafted features to detect tube-like structures such as vessels. However, those methods require complex pipelines involving several hyper-parameters and design choices rendering the procedure sensitive, dataset-specific, and not generalizable. We propose a self-supervised method with a limited number of hyperparameters that is generalizable across modalities. Our method uses tube-like structure properties, such as connectivity, profile consistency, and bifurcation, to introduce inductive bias into a learning algorithm. To model those properties, we generate a vector field that we refer to as a flow. Our experiments on various public datasets in 2D and 3D show that our method performs better than unsupervised methods while learning useful transferable features from unlabeled data. Unlike generic self-supervised methods, the learned features learn vessel-relevant features that are transferable for supervised approaches, which is essential when the number of annotated data is limited.
Abstract
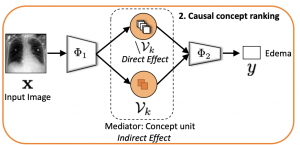
Model explainability is essential for the creation of trustworthy Machine Learning models in healthcare. An ideal explanation resembles the decision-making process of a domain expert and is expressed using concepts or terminology that is meaningful to the clinicians. To provide such an explanation, we first associate the hidden units of a Black-box to clinically relevant concepts. We take advantage of radiology reports accompanying the chest X-ray images to define concepts. We discover sparse associations between concepts and hidden units using a linear sparse logistic regression. To ensure that the identified units truly influence the classifier’s outcome, we adopt tools from Causal Inference literature and, more specifically, mediation analysis through counterfactual interventions. Finally, we construct a low-depth decision tree to translate all the discovered concepts into a straightforward decision rule, expressed to the radiologist. We evaluated our approach on a large chest x-ray dataset, where our model produces a global explanation consistent with clinical knowledge.
Empowering Variational Inference with Predictive Features: Application to Disease Suptypying
A. Saeedi, P. Yadollahpour, S. Singla, W. Wells, F. Sciurba, K. Batmanghelich
Machine Learning in Healthcare Conference (MLHC 21)
[pdf]
Abstract
Generative models, such as the probabilistic topic model, have been widely deployed for various applications in the healthcare domain, such as learning disease or tissue subtypes. However, learning the parameters of such models is usually an ill-posed problem and may lose valuable information about disease severity. A common approach is to add a discriminative loss to the generative model’s learning loss; finding a balance between two losses is not straightforward. We propose an alternative way in this paper. We use distribution embedding to construct patient-level representation, which is usually more discriminative than the posterior parameters. We view the patient-level representation as an external covariate. Then, we use the external covariates to inform the posterior of our generative model. Effectively, we enforce the generative model’s approximate posterior to reside in the subspace of the discriminative covariates. We illustrate this method’s application on a large-scale lung CT study of Chronic Obstructive Pulmonary Disease (COPD), a highly heterogeneous disease. We aim at identifying tissue subtypes by using a variant of a topic model as a generative model. We evaluate the patient representation, the resulting topics on the patient- and population-level. We also show that some of the discovered subtypes are correlated with genetic measurements, suggesting that the identified subtypes characterize the disease’s underlying etiology.
Abstract
To develop and evaluate a deep learning (DL) approach to extract rich information from High-Resolution Computed Tomography (HRCT) of patients with chronic obstructive pulmonary disease (COPD). We develop a DL based model to learn a compact representation of a subject, which is predictive of COPD physiologic severity and other outcomes. Our DL model learned: 1) to extract informative regional image features from HRCT, 2) to adaptively weight these features and form an aggregate patient representation, and finally 3) to predict several COPD outcomes. The adaptive weights correspond to the regional lung contribution to the disease. We evaluate the model on 10,300 participants from the COPDGene cohort. Our model was strongly predictive of spirometric obstruction (r2= 0.67) and grouped 65.4% of subjects correctly and 89.1% within one stage of their GOLD severity stage. Our model achieved an accuracy of 41.7% and 52.8% in stratifying the population-based on centrilobular (5-grade) and paraseptal (3-grade) emphysema severity score, respectively. For predicting future exacerbation, combining subjects’ representations from our model with their past exacerbation histories achieved an accuracy of 80.8% (area under the ROC curve of 0.73). For all-cause mortality, in Cox-regression analysis, we outperformed the BODE index improving the concordance metric (ours: 0.61 vs. BODE: 0.56). Our model independently predicted spirometric obstruction, emphysema severity, exacerbation risk, and mortality from CT imaging alone. This method has potential applicability in both research and clinical practice.
Abstract
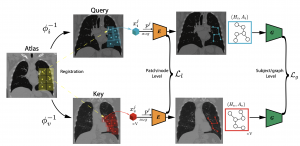
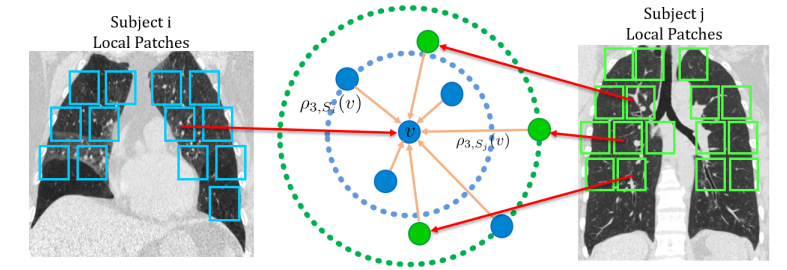
Supervised learning method requires a large volume of annotated datasets. Collecting such datasets is time-consuming and expensive. Until now, very few annotated COVID-19 imaging datasets are available. Although self-supervised learning enables us to bootstrap the training by exploiting unlabeled data, the generic self-supervised methods for natural images do not sufficiently incorporate the context. For medical images, a desirable method should be sensitive enough to detect deviation from normal-appearing tissue of each anatomical region; here, anatomy is the context. We introduce a novel approach with two levels of self-supervised representation learning objectives: one on the regional anatomical level and another on the patient-level. We use graph neural networks to incorporate the relationship between different anatomical regions. The structure of the graph is informed by anatomical correspondences between each patient and an anatomical atlas. In addition, the graph representation has the advantage of handling any arbitrarily sized image in full resolution. Experiments on large-scale Computer Tomography (CT) datasets of lung images show that our approach compares favorably to baseline methods that do not account for the context. We use the learned embedding to quantify the clinical progression of COVID-19 and show that our method generalizes well to COVID-19 patients from different hospitals. Qualitative results suggest that our model can identify clinically relevant regions in the images.
2020
Abstract
To achieve a holistic view of the underlying mechanisms of human diseases, the biomedical research community is moving toward harvesting retrospective data available in Electronic Healthcare Records (EHRs). The first step for causal understanding is to perform association tests between types of potentially high-dimensional biomedical data, such as genetic, blood biomarkers, and imaging data. To obtain a reasonable power, current methods require a substantial sample size of individuals with both data modalities. This prevents researchers from using much larger EHR samples that include individuals with at least one data type, limits the power of the association test, and may result in a higher false discovery rate. We present a new method called the Semi-paired Association Test (SAT) that makes use of both paired and unpaired data. In contrast to classical approaches, incorporating unpaired data allows SAT to produce better control of false discovery and, under some conditions, improve the association test power. We study the properties of SAT theoretically and empirically, through simulations and application to real studies in the context of Chronic Obstructive Pulmonary Disease. Our method identifies an association between the high-dimensional characterization of Computed Tomography (CT) chest images and blood biomarkers as well as the expression of dozens of genes involved in the immune system.
Label-Noise Robust Domain Adaptation
Xiyu Yu, Tongliang Liu, Mingming Gong, Kun Zhang, Kayhan Batmanghelich, Dacheng Tao
ICML, 2020
[pdf]
Abstract
Domain adaptation aims to correct the classifiers when faced with a distribution shift between the source (training) and target (test) domains. State-of-the-art domain adaptation methods make use of deep networks to extract domain-invariant representations. However, existing methods assume that all the instances in the source domain are correctly labeled; while in reality, it is unsurprising that we may obtain a source domain with noisy labels. In this paper, we are the first to comprehensively investigate how the label-noise could adversely affect existing domain adaptation methods in various scenarios. Further, we theoretically prove that there exists a method that can essentially reduce the side-effect of noisy source labels in domain adaptation. Specifically, focusing on the generalized target shift scenario, where both label distribution $P_Y$ and the class-conditional distribution $P_{X|Y}$ can change, we discover that the denoising Conditional Invariant Component (DCIC) framework can provably ensure (1) extracting invariant representations given examples with noisy labels in the source domain and unlabeled examples in the target domain and (2) estimating the label distribution in the target domain with no bias. Experimental results on both synthetic and real-world data verify the effectiveness of the proposed method.
Abstract
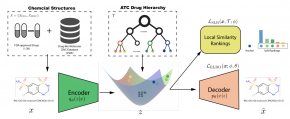
Learning accurate drug representation is essential for tasks such as computational drug repositioning and prediction of drug side-effects. A drug hierarchy is a valuable source that encodes human knowledge of drug relations in a tree-like structure where drugs that act on the same organs, treat the same disease, or bind to the same biological target are grouped together. However, its utility in learning drug representations has not yet been explored, and currently described drug representations cannot place novel molecules in a drug hierarchy. Here, we develop a semi-supervised drug embedding that incorporates two sources of information: (1) underlying chemical grammar that is inferred from molecular structures of drugs and drug-like molecules (unsupervised), and (2) hierarchical relations that are encoded in an expert-crafted hierarchy of approved drugs (supervised). We use the Variational Auto-Encoder (VAE) framework to encode the chemical structures of molecules and use the knowledge-based drug-drug similarity to induce the clustering of drugs in hyperbolic space. The hyperbolic space is amenable for encoding hierarchical concepts. Both quantitative and qualitative results support that the learned drug embedding can accurately reproduce the chemical structure and induce the hierarchical relations among drugs. Furthermore, our approach can infer the pharmacological properties of novel molecules by retrieving similar drugs from the embedding space. We demonstrate that the learned drug embedding can be used to find new uses for existing drugs and to discover side-effects. We show that it significantly outperforms baselines in both tasks.
3D-BoxSup: Positive-Unlabeled Learning of Brain Tumor Segmentation Networks From 3D Bounding Boxes
Yanwu Xu, Mingming Gong, Junxiang Chen, Ziye Chen, Kayhan Batmanghelich
Frontiers in Neuroscience, 2020
[pdf]
Abstract
Accurate segmentation is an essential task when working with medical images. Recently, deep convolutional neural networks achieved state-of-the-art performance for many segmentation benchmarks. Regardless of the network architecture, the deep learning-based segmentation methods view the segmentation problem as a supervised task that requires a relatively large number of annotated images. Acquiring a large number of annotated medical images is time-consuming, and high-quality segmented images (i.e., strong labels) crafted by human experts are expensive. In this paper, we have proposed a method that achieves competitive accuracy from a “weakly annotated” image where the weak annotation is obtained via a 3D bounding box denoting an object of interest. Our method, called “3D-BoxSup,” employs a positive-unlabeled learning framework to learn segmentation masks from 3D bounding boxes. Specially, we consider the pixels outside of the bounding box as positively labeled data and the pixels inside the bounding box as unlabeled data. Our method can suppress the negative effects of pixels residing between the true segmentation mask and the 3D bounding box and produce accurate segmentation masks. We applied our method to segment a brain tumor. The experimental results on the BraTS 2017 dataset (Menze et al., 2015; Bakas et al., 2017a,b,c) have demonstrated the effectiveness of our method.
Human-Machine Collaboration for Medical Image Segmentation
Mahdyar Ravanbakhsh, Vadim Tschernezki, Felix Last, Tassilo Klein, Kayhan Batmanghelich, Volker Tresp, Moin Nabi
IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), 2020
[pdf]
Abstract
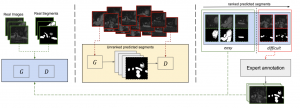
Image segmentation is a ubiquitous step in almost any medical image study. Deep learning-based approaches achieve state-of-the-art in the majority of image segmentation benchmarks. However, end-to-end training of such models requires sufficient annotation. In this paper, we propose a method based on conditional Generative Adversarial Network (cGAN) to address segmentation in semi-supervised setup and in a human-in-the-loop fashion. More specifically, we use the generator in the GAN to synthesize segmentations on unlabeled data and use the discriminator to identify unreliable slices for which expert annotation is required. The quantitative results on a conventional standard benchmark show that our method is comparable with the state-of-the-art fully supervised methods in slice-level evaluation, despite of requiring far less annotated data.
Abstract
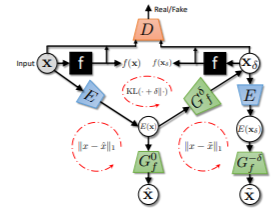
As machine learning methods see greater adoption and implementation in high stakes applications such as medical image diagnosis, the need for model interpretability and explanation has become more critical. Classical approaches that assess feature importance (eg saliency maps) do not explain how and why a particular region of an image is relevant to the prediction. We propose a method that explains the outcome of a classification black-box by gradually exaggerating the semantic effect of a given class. Given a query input to a classifier, our method produces a progressive set of plausible variations of that query, which gradually changes the posterior probability from its original class to its negation. These counter-factually generated samples preserve features unrelated to the classification decision, such that a user can employ our method as a “tuning knob” to traverse a data manifold while crossing the decision boundary. Our method is model agnostic and only requires the output value and gradient of the predictor with respect to its input.
Generative-Discriminative Complementary Learning
Yanwu Xu, Mingming Gong, Junxiang Chen, Tongliang Liu, Kun Zhang, Kayhan Batmanghelich
Proceeding of Thirty-Fourth AAAI Conference on Artificial Intelligence (AAAI-20)
[pdf]
Abstract
The majority of state-of-the-art deep learning methods are discriminative approaches, which model the conditional distribution of labels given input features. The success of such approaches heavily depends on high-quality labeled instances, which are not easy to obtain, especially as the number of candidate classes increases. In this paper, we study the complementary learning problem. Unlike ordinary labels, complementary labels are easy to obtain because an annotator only needs to provide a yes/no answer to a randomly chosen candidate class for each instance. We propose a generative-discriminative complementary learning method that estimates the ordinary labels by modeling both the conditional (discriminative) and instance (generative) distributions. Our method, we call Complementary Conditional GAN (CCGAN), improves the accuracy of predicting ordinary labels and is able to generate high-quality instances in spite of weak supervision. In addition to the extensive empirical studies, we also theoretically show that our model can retrieve the true conditional distribution from the complementarily-labeled data.
Abstract
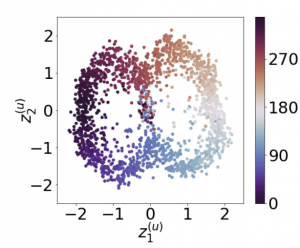
Recently, researches related to unsupervised disentanglement learning with deep generative models have gained substantial popularity. However, without introducing supervision, there is no guarantee that the factors of interest can be successfully recovered. In this paper, we propose a setting where the user introduces weak supervision by providing similarities between instances based on a factor to be disentangled. The similarity is provided as either a discrete (yes/no) or real-valued label describing whether a pair of instances are similar or not. We propose a new method for weakly supervised disentanglement of latent variables within the framework of Variational Autoencoder. Experimental results demonstrate that utilizing weak supervision improves the performance of the disentanglement method substantially.
2019
Abstract
Unsupervised domain mapping aims to learn a function to translate domain X to Y by a function GXY in the absence of paired examples. Finding the optimal GXY without paired data is an ill-posed problem, so appropriate constraints are required to obtain reasonable solutions. One of the most prominent constraints is cycle consistency, which enforces the translated image by GXY to be translated back to the input image by an inverse mapping GYX. While cycle consistency requires the simultaneous training of GXY and GY X, recent studies have shown that one-sided domain mapping can be achieved by preserving pairwise distances between images. Although cycle consistency and distance preservation successfully constrain the solution space, they overlook the special properties that simple geometric transformations do not change the semantic structure of images. Based on this special property, we develop a geometry-consistent generative adversarial network (GcGAN), which enables one-sided unsupervised domain mapping. GcGAN takes the original image and its counterpart image transformed by a predefined geometric transformation as inputs and generates two images in the new domain coupled with the corresponding geometry-consistency constraint. The geometry-consistency constraint reduces the space of possible solutions while keeping the correct solutions in the search space. Quantitative and qualitative comparisons with the baseline (GAN alone) and the state-of-the-art methods including CycleGAN and DistanceGAN demonstrate the effectiveness of our method.
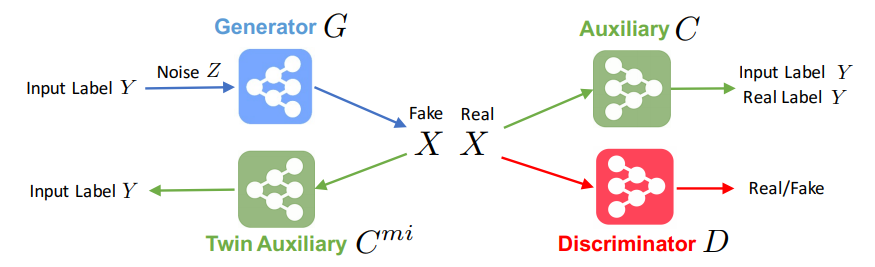
Abstract
Conditional generative models enjoy remarkable progress over the past few years. One of the popular conditional models is Auxiliary Classifier GAN (AC-GAN), which generates highly discriminative images by extending the loss function of GAN with an auxiliary classifier. However, the diversity of the generated samples by AC-GAN tends to decrease as the number of classes increases, hence limiting its power on large-scale data. In this paper, we identify the source of the low diversity issue theoretically and propose a practical solution to solve the problem. We show that the auxiliary classifier in AC-GAN imposes perfect separability, which is disadvantageous when the supports of the class distributions have significant overlap. To address the issue, we propose Twin Auxiliary Classifiers Generative Adversarial Net (TAC-GAN) that further benefits from a new player that interacts with other players (the generator and the discriminator) in GAN. Theoretically, we demonstrate that TAC-GAN can effectively minimize the divergence between the generated and real-data distributions. Extensive experimental results show that our
TAC-GAN can successfully replicate the true data distributions on simulated data, and significantly improves the diversity of class-conditional image generation on real datasets.
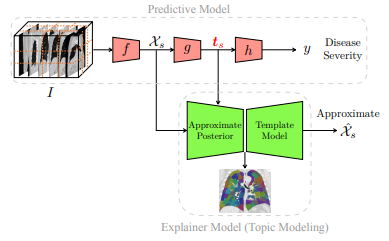
Generative Interpretability: Application in Disease Subtyping
P. Yadollahpour, A. Saeedi, S. Singla, F. C. Sciurba, K. Batmanghelich
Pre-print (Report)
[pdf]
Abstract
We present a probabilistic approach to characterize heterogeneous disease in a way that is reflective of disease severity. In many diseases, multiple subtypes of disease present simultaneously in each patient. Generative models provide a flexible and readily explainable framework to discover disease subtypes from imaging data. However, discovering local image descriptors of each subtype in a fully unsupervised way is an ill-posed problem and may result in loss of valuable information about disease severity. Although supervised approaches, and more recently deep learning methods, have achieved state-of-the-art performance for predicting clinical variables relevant to diagnosis, interpreting those models is a crucial yet challenging task. In this paper, we propose a method that aims to achieve the best of both worlds, namely we maintain the predictive power of supervised methods and the interpretability of probabilistic methods. Taking advantage of recent progress in deep learning, we propose to incorporate the discriminative information extracted by the predictive model into the posterior distribution over the latent variables of the generative model. Hence, one can view the generative model as a template for interpretation of a discriminative method in a clinically meaningful way. We illustrate an application of this method on a large-scale lung CT study of Chronic Obstructive Pulmonary Disease (COPD), which is a highly heterogeneous disease. As our experiments show, our interpretable model does not compromise the prediction of the relevant clinical variables, unlike purely unsupervised methods. We also show that some of the discovered subtypes are correlated with genetic measurements suggesting that the discovered subtypes characterize the underlying etiology of the disease.
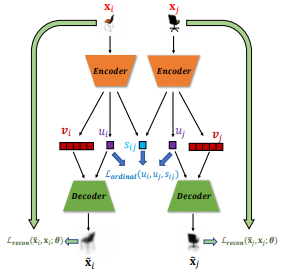
Abstract
Recent work by Locatello et al. (2018) has shown that an inductive bias is required to disentangle factors of interest in Variational Autoencoder (VAE). Motivated by a real-world problem, we propose a setting where such bias is introduced by providing pairwise ordinal comparisons between instances, based on the desired factor to be disentangled. For example, a doctor compares pairs of patients based on the level of severity of their illnesses, and the desired factor is a quantitive level of the disease severity. In a real-world application, the pairwise comparisons are usually noisy. Our method, Robust Ordinal VAE (ROVAE), incorporates the noisy pairwise ordinal comparisons in the disentanglement task. We introduce non-negative random variables in ROVAE, such that it can automatically determine whether each pairwise ordinal comparison is trustworthy and ignore the noisy comparisons. Experimental results demonstrate that ROVAE outperforms existing methods and is more robust to noisy pairwise comparisons in both benchmark datasets and a real-world application.
2018
Subject2Vec: Generative-Discriminative Approach from a Set of Image Patches to a Vector
S. Singla, M. Gong, S. Ravanbakhsh, F. Sciurba, B. Poczos, K. N. Batmanghelich
Medical Image Computing & Computer-Assisted Intervention
[pdf]
Abstract
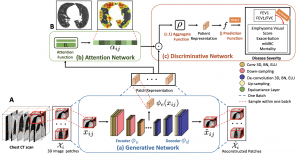
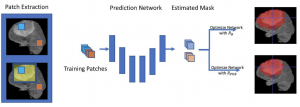
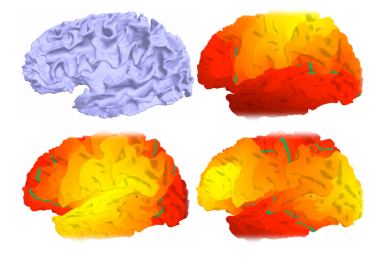
We propose an attention-based method that aggregates local image features to a subject-level representation for predicting disease severity. In contrast to classical deep learning that requires a fixed dimensional input, our method operates on a set of image patches; hence it can accommodate variable length input image without image resizing. The model learns a clinically interpretable subject-level representation that is reflective of the disease severity. Our model consists of three mutually dependent modules which regulate each other: (1) a discriminative network that learns a fixed-length representation from local features and maps them to disease severity; (2) an attention mechanism that provides interpretability by focusing on the areas of the anatomy that contribute the most to the prediction task; and (3) a generative network that encourages the diversity of the local latent features. The generative term ensures that the attention weights are non-degenerate while maintaining the relevance of the local regions to the disease severity. We train our model end-to-end in the context of a large-scale lung CT study of Chronic Obstructive Pulmonary Disease (COPD). Our model gives state-of-the-art performance in predicting clinical measures of severity for COPD. The distribution of the attention provides the regional relevance of lung tissue to the clinical measurements.
A structural equation model for imaging genetics using spatial transcriptomics
S. M. H. Huisman, A. Mahfouz, K. Batmanghelich, B. P. F. Lelieveldt, M. J. T. Reinders
Brain Informatics
[pdf]
Abstract
Imaging genetics deals with relationships between genetic variation and imaging variables, often in a disease context. The complex relationships between brain volumes and genetic variants have been explored with both dimension reduction methods and model-based approaches. However, these models usually do not make use of the extensive knowledge of the spatio-anatomical patterns of gene activity. We present a method for integrating genetic markers (single nucleotide polymorphisms) and imaging features, which is based on a causal model and, at the same time, uses the power of dimension reduction. We use structural equation models to find latent variables that explain brain volume changes in a disease context, and which are in turn affected by genetic variants. We make use of publicly available spatial transcriptome data from the Allen Human Brain Atlas to specify the model structure, which reduces noise and improves interpretability. The model is tested in a simulation setting and applied on a case study of the Alzheimer’s Disease Neuroimaging Initiative.
Causal Generative Domain Adaptation Networks
M. Gong, K. Zhang, B. Huang, C. Glymour, D. Tao, K. Batmanghelich
Preprint: arXiv:1804.04333
[pdf]
Abstract
We propose a new generative model for domain adaptation, in which training data (source domain) and test data (target domain) come from different distributions. An essential problem in domain adaptation is to understand how the distribution shifts across domains. For this purpose, we propose a generative domain adaptation network to understand and identify the domain changes, which enables the generation of new domains. In addition, focusing on single domain adaptation, we demonstrate how our model recovers the joint distribution on the target domain from unlabeled target domain data by transferring valuable information between domains. Finally, to improve transfer efficiency, we build a causal generative domain adaptation network by decomposing the joint distribution of features and labels into a series of causal modules according to a causal model. Due to the modularity property of a causal model, we can improve the identification of distribution changes by modeling each causal modules separately. With the proposed adaptation networks, the predictive model on the target domain can be easily trained on data sampled from the learned networks. We demonstrate the efficacy of our method on both synthetic and real data experiments.
Deep Diffeomorphic Normalizing Flows
H. Salman, P. Yadollahpour, T. Fletcher, K. Batmanghelich
Preprint: arXiv:1810.03256
[pdf]
Abstract
The Normalizing Flow (NF) models a general probability density by estimating an invertible transformation applied on samples drawn from a known distribution. We introduce a new type of NF, called Deep Diffeomorphic Normalizing Flow (DDNF). A diffeomorphic flow is an invertible function where both the function and its inverse are smooth. We construct the flow using an ordinary differential equation (ODE) governed by a time-varying smooth vector field. We use a neural network to parametrize the smooth vector field and a recursive neural network (RNN) for approximating the solution of the ODE. Each cell in the RNN is a residual network implementing one Euler integration step. The architecture of our flow enables efficient likelihood evaluation, straightforward flow inversion, and results in highly flexible density estimation. An end-to-end trained DDNF achieves competitive results with state-of-the-art methods on a suite of density estimation and variational inference tasks. Finally, our method brings concepts from Riemannian geometry that, we believe, can open a new research direction for neural density estimation.
Abstract
In this paper, we study the mixture proportion estimation (MPE) problem in a new setting: given samples from the mixture and the component distributions, we identify the proportions of the components in the mixture distribution. To address this problem, we make use of a linear independence assumption, ie, the component distributions are independent from each other, which is much weaker than assumptions exploited in the previous MPE methods. Based on this assumption, we propose a method (1) that uniquely identifies the mixture proportions,(2) whose output provably converges to the optimal solution, and (3) that is computationally efficient. We show the superiority of the proposed method over the state-of-the-art methods in two applications including learning with label noise and semi-supervised learning on both synthetic and real-world datasets.
Abstract
Monocular depth estimation, which plays a crucial role in understanding 3D scene geometry, is an ill-posed problem. Recent methods have gained significant improvement by exploring image-level information and hierarchical features from deep convolutional neural networks (DCNNs). These methods model depth estimation as a regression problem and train the regression networks by minimizing mean squared error, which suffers from slow convergence and unsatisfactory local solutions. Besides, existing depth estimation networks employ repeated spatial pooling operations, resulting in undesirable low-resolution feature maps. To obtain high-resolution depth maps, skip-connections or multilayer deconvolution networks are required, which complicates network training and consumes much more computations. To eliminate or at least largely reduce these problems, we introduce a spacing-increasing discretization (SID) strategy to discretize depth and recast depth network learning as an ordinal regression problem. By training the network using an ordinary regression loss, our method achieves much higher accuracy and faster convergence in synch. Furthermore, we adopt a multi-scale network structure which avoids unnecessary spatial pooling and captures multi-scale information in parallel. The proposed deep ordinal regression network (DORN) achieves state-of-the-art results on three challenging benchmarks, ie, KITTI, Make3D, and NYU Depth v2, and outperforms existing methods by a large margin.
Textured Graph-Based Model of the Lungs: Application on Tuberculosis Type Classification and Multi-drug Resistance Detection
Y.D. Cid, K. Batmanghelich, H. Müller
International Conference of the Cross-Language Evaluation Forum for European Languages
[pdf]
Abstract
Tuberculosis (TB) remains a leading cause of death worldwide. Two main challenges when assessing computed tomography scans of TB patients are detecting multi-drug resistance and differentiating TB types. In this article we model the lungs as a graph entity where nodes represent anatomical lung regions and edges encode interactions between them. This graph is able to characterize the texture distribution along the lungs, making it suitable for describing patients with different TB types. In 2017, the ImageCLEF benchmark proposed a task based on computed tomography volumes of patients with TB. This task was divided into two subtasks: multi-drug resistance prediction, and TB type classification. The participation in this task showed the strength of our model, leading to best results in the competition for multi-drug resistance detection (AUC = 0.5825) and good results in the TB type classification (Cohen’s Kappa coefficient = 0.1623).
2017
Transformations Based on Continuous Piecewise-Affine Velocity Fields
O. Freifeld, S. Hauberg, J. Fisher III, N. Batmanghelich
IEEE Transactions on Pattern Analysis and Machine Intelligence
[pdf]
Abstract
We propose novel finite-dimensional spaces of well-behaved Rn → Rn transformations. The latter are obtained by (fast and highly-accurate) integration of continuous piecewise-affine velocity fields. The proposed method is simple yet highly expressive, effortlessly handles optional constraints (e.g., volume preservation and/or boundary conditions), and supports convenient modeling choices such as smoothing priors and coarse-to-fine analysis. Importantly, the proposed approach, partly due to its rapid likelihood evaluations and partly due to its other properties, facilitates tractable inference over rich transformation spaces, including using Markov-Chain Monte-Carlo methods. Its applications include, but are not limited to: monotonic regression (more generally, optimization over monotonic functions); modeling cumulative distribution functions or histograms; time-warping; image warping; image registration; real-time diffeomorphic image editing; data augmentation for image classifiers. Our GPU-based code is publicly available.
A Likelihood-Free Approach for Characterizing Heterogeneous Diseases in Large-Scale Studies
J. Schabdach, S. Wells, M. Cho, N. Batmanghelich
Information Processing in Medical Imaging (IPMI)
[pdf]
Abstract
We propose a non-parametric approach for characterizing heterogeneous diseases in large-scale studies. We target diseases where multiple types of pathology present simultaneously in each subject and a more severe disease manifests as a higher level of tissue destruction. For each subject, we model the collection of local image descriptors as samples generated by an unknown subject-specic probability density. Instead of approximating the probability density via a parametric family, we propose to side step the parametric inference by directly estimating the divergence between subject densities. Our method maps the collection of local image descriptors to a signature vector that is used to predict a clinical measurement. We are able to interpret the prediction of the clinical variable in the population and individual levels by carefully studying the divergences. We illustrate an application this method on simulated data as well as on a large-scale lung CT study of Chronic Obstructive Pulmonary Disease (COPD). Our approach outperforms classical methods on both simulated and COPD data and demonstrates the state-of-the-art prediction on an important physiologic measure of airflow (the forced respiratory volume in one second, FEV1).
2016
Unsupervised Discovery of Emphysema Subtypes in a Large Clinical Cohort
P. Binder N. Batmanghelich, R. J. Estepar, P. Golland
7th International Workshop on Machine Learning in Medical Imaging (MLMI)
[pdf]
Abstract
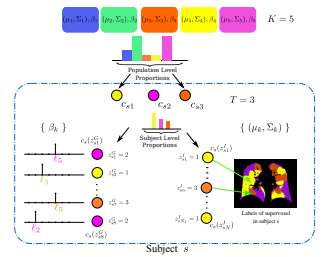
Emphysema is one of the hallmarks of Chronic Obstructive Pulmonary Disorder (COPD), a devastating lung disease often caused by smoking. Emphysema appears on Computed Tomography (CT) scans as a variety of textures that correlate with disease subtypes. It has been shown that the disease subtypes and textures are linked to physiological indicators and prognosis, although neither is well characterized clinically. Most previous computational approaches to modeling emphysema imaging data have focused on supervised classification of lung textures in patches of CT scans. In this work, we describe a generative model that jointly captures heterogeneity of disease subtypes and of the patient population. We also describe a corresponding inference algorithm that simultaneously discovers disease subtypes and population structure in an unsupervised manner. This approach enables us to create image-based descriptors of emphysema beyond those that can be identified through manual labeling of currently defined phenotypes. By applying the resulting algorithm to a large data set, we identify groups of patients and disease subtypes that correlate with distinct physiological indicators.
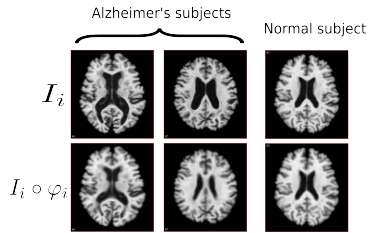
Probabilistic Modeling of Imaging, Genetics and the Diagnosis
K.N. Batmanghelich, A. Dalca, G. Quon, M. Sabuncu, P. Golland
IEEE Trans Med Imaging
[pdf]
Abstract
We propose a unified Bayesian framework for detecting genetic variants associated with disease by exploiting imagebased features as an intermediate phenotype. The use of imaging data for examining genetic associations promises new directions of analysis, but currently the most widely used methods make sub-optimal use of the richness that these data types can offer. Currently, image features are most commonly selected based on their relevance to the disease phenotype. Then, in a separate step, a set of genetic variants is identified to explain theselected features. In contrast, our method performs these tasks simultaneously in order to jointly exploit information in both data types. The analysis yields probabilistic measures of clinical relevance for both imaging and genetic markers. We derive an efficient approximate inference algorithm that handles the high dimensionality of image and genetic data. We evaluate the algorithm on synthetic data and demonstrate that it outperforms traditional models. We also illustrate our method on Alzheimer’s Disease Neuroimaging Initiative data.
Nonparametric Spherical Topic Modeling with Word Embeddings
N. Batmanghelich, A. Saeediy, K. Narasimhan, S. Gershman
Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics (ACL)
[pdf]
Abstract
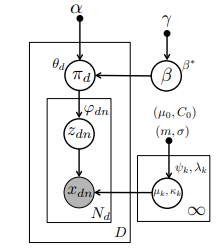
Traditional topic models do not account for semantic regularities in language. Recent distributional representations of words exhibit semantic consistency over directional metrics such as cosine similarity. However, neither categorical nor Gaussian observational distributions used in existing topic models are appropriate to leverage such correlations. In this paper, we propose to use the von Mises-Fisher distribution to model the density of words over a unit sphere. Such a representation is well-suited for directional data. We use a Hierarchical Dirichlet Process for our base topic model and propose an efficient inference algorithm based on Stochastic Variational Inference. This model enables us to naturally exploit the semantic structures of word embeddings while flexibly discovering the number of topics. Experiments demonstrate that our method outperforms competitive approaches in terms of topic coherence on two different text corpora while offering efficient inference.
Inferring Disease Status by non-Parametric Probabilistic Embedding
N. Batmanghelich, A. Saeedi, R. J. Estepar, M. Cho, S. Wells
Workshop on Medical Computer Vision: Algorithms for Big Data (MCV)
[pdf]
Abstract
Computing similarity between all pairs of patients in a dataset enables us to group the subjects into disease subtypes and infer their disease status. However, robust and efficient computation of pairwise similarity is challenging task for large-scale medical image datasets. We specifically target diseases where multiple subtypes of pathology present simultaneously, rendering the denition of the similarity a difficult task. To define pairwise patient similarity, we characterize each subject by a probability distribution that generates its local image descriptors. We adopt a notion of affinity between probability distributions which lends itself to similarity between subjects. Instead of approximating the distributions by a parametric family, we propose to compute the affinity measure indirectly using an approximate nearest neighbor estimator. Computing pairwise similarities enables us to embed the entire patient population into a lower dimensional manifold, mapping each subject from high-dimensional image space to an informative low-dimensional representation. We validate our method on a large-scale lung CT scan study and demonstrate the state-of-the-art prediction on an important physiologic measure of airflow (the forced expiratory volume in one second, FEV1) in addition to a 5-category clinical rating (so-called GOLD score).
2015
Highly-Expressive Spaces of Well- Behaved Transformations: Keeping It Simple
O. Freifeld, S. Hauberg, N. Batmanghelich
Proceedings of the IEEE International Conference on Computer Vision (ICCV)
[pdf]
Abstract
Generative Method to Discover Genetically Driven Image Biomarkers
K.N. Batmanghelich*, A. Saeedi*, M. Cho, R. Jose, P. Golland
In Proc. IPMI: International Conference on Information Processing and Medical Imaging
[pdf]
Abstract
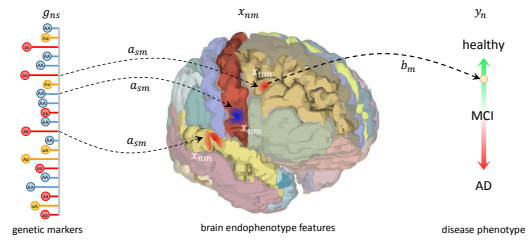
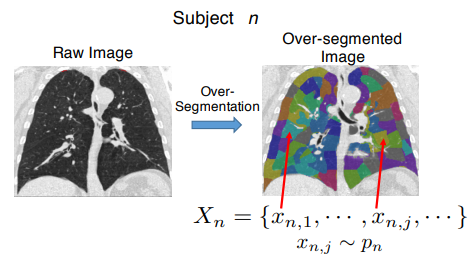
We present a generative probabilistic approach to discovery of disease subtypes determined by the genetic code. In many diseases, multiple types of pathology may present simultaneously in a patient, making quantification of the disease challenging. Our method seeks common co-occurring image and genetic patterns in a population as a way to model these two different data types jointly. We assume that each patient is a mixture of multiple disease subtypes and use the joint generative model of image and genetic markers to identify disease subtypes guided by known genetic influences. Our model is based on a variant of the so-called topic models that uncover the latent structure in a collection of data. We derive an efficient variational inference algorithm to extract patterns of co-occurrence and to quantify the presence of a heterogeneous disease in each patient. We evaluate the method on simulated data and illustrate its use in the context of Chronic Obstructive Pulmonary Disease (COPD) to characterize the relationship between image and genetic signatures of the COPD subtypes in a large patient cohort.
2014
Spherical Topic Models for Imaging Phenotype Discovery in Genetic Studies
K.N. Batmanghelich, M. Cho, R. Jose, P. Golland
In Proc. BAMBI: Workshop on Bayesian and Graphical Models for Biomedical imaging, International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI)
[pdf]
Abstract
In this paper, we use Spherical Topic Models to discover the latent structure of lung disease. This method can be widely employed when a measurement for each subject is provided as a normalized histogram of relevant features. In this paper, the resulting descriptors are used as phenotypes to identify genetic markers associated with the Chronic Obstructive Pulmonary Disease (COPD). Features extracted from images capture the heterogeneity of the disease and therefore promise to improve detection of relevant genetic variants in Genome Wide Association Studies (GWAS). Our generative model is based on normalized histograms of image intensity of each subject and it can be readily extended to other forms of features as long as they are provided as normalized histograms. The resulting algorithm represents the intensity distribution as a combination of meaningful latent factors and mixing coefficients that can be used for genetic association analysis. This approach is motivated by a clinical hypothesis that COPD symptoms are caused by multiple coexisting disease processes. Our experiments show that the new features enhance the previously detected signal on chromosome 15 with respect to standard respiratory and imaging measurements.
Diversifying Sparsity Using Variational Determinantal Point Processes
K.N. Batmanghelich, G. Quon, A. Kuleza, M. Kellis, P. Golland, L. Bornn
In Proc. ArXiv
[pdf]
Abstract
We propose a novel diverse feature selection method based on determinantal point processes (DPPs). Our model enables one to flexibly define diversity based on the covariance of features (similar to orthogonal matching pursuit) or alternatively based on side information. We introduce our approach in the context of Bayesian sparse regression, employing a DPP as a variational approximation to the true spike and slab posterior distribution. We subsequently show how this variational DPP approximation generalizes and extends mean-field approximation, and can be learned efficiently by exploiting the fast sampling properties of DPPs. Our motivating application comes from bioinformatics, where we aim to identify a diverse set of genes whose expression profiles predict a tumor type where the diversity is defined with respect to a gene-gene interaction network. We also explore an application in spatial statistics. In both cases, we demonstrate that the proposed method yields significantly more diverse feature sets than classic sparse methods, without compromising accuracy.
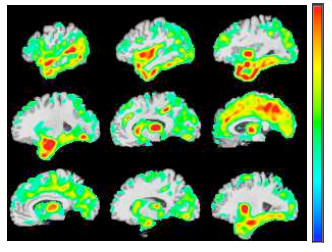
BrainPrint in the Computer-Aided Diagnosis of Alzheimer’s Disease
C. Wachinger, K. Batmanghelich, P. Golland, M. Reuter
Challenge on Computer-Aided Diagnosis of Dementia, MICCAI, 2014
[pdf]
Abstract
We investigate the potential of shape information in assisting the computer-aided diagnosis of Alzheimer’s disease and its prodromal stage of mild cognitive impairment. We employ BrainPrint to obtain an extensive characterization of the shape of brain structures. BrainPrint captures shape information of an ensemble of cortical and subcortical structures by solving the 2D and 3D Laplace-Beltrami operator on triangular and tetrahedral meshes. From the shape descriptor, we derive features for the classi cation by computing lateral shape di erences and the projection on the principal component. Volume and thickness measurements from FreeSurfer complement the shape features in our model. We use the generalized linear model with a multinomial link function for the classification. Next to manual model selection, we employ the elastic-net regularizer and stepwise model selection with the Akaike information criterion. Training is performed on data provided by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) and testing on the data provided by the challenge. The approach runs fully automatically.
2013
Joint Modeling of Imaging and Genetics
K.N. Batmanghelich, A.V. Dalca, M.R. Sabuncu, P. Golland
In Proc. IPMI: International Conference on Information Processing and Medical Imaging
[pdf]
Abstract
We propose a unified Bayesian framework for detecting genetic variants associated with a disease while exploiting image-based features as an intermediate phenotype. Traditionally, imaging genetics methods comprise two separate steps. First, image features are selected based on their relevance to the disease phenotype. Second, a set of genetic variants are identified to explain the selected features. In contrast, our method performs these tasks simultaneously to ultimately assign probabilistic measures of relevance to both genetic and imaging markers. We derive an efficient approximate inference algorithm that handles high dimensionality of imaging genetic data. We evaluate the algorithm on synthetic data and show that it outperforms traditional models. We also illustrate the application of the method on ADNI data.
2012
Dominant Component Analysis of Electro- physiological Connectivity Network
Y. Ghanbari, L. Bloy, N. Batmanghelich, R. Verma
International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI)
[pdf]
Abstract
Connectivity matrices obtained from various modalities (DTI, MEG and fMRI) provide a unique insight into brain processes. Their high dimensionality necessitates the development of methods for population-based statistics, in the face of small sample sizes. In this paper, we present such a method applicable to functional connectivity networks, based on identifying the basis of dominant connectivity components that characterize the patterns of brain pathology and population variation. Projection of individual connectivity matrices into this basis allows for dimensionality reduction, facilitating subsequent statistical analysis. We find dominant components for a collection of connectivity matrices by using the projective non-negative component analysis technique which ensures that the components have non-negative elements and are non-negatively combined to obtain individual subject networks, facilitating interpretation. We demonstrate the feasibility of our novel framework by applying it to simulated connectivity matrices as well as to a clinical study using connectivity matrices derived from resting state magnetoencephalography (MEG) data in a population of subjects diagnosed with autism spectrum disorder (ASD).
An integrated Framework for High Angular Resolution Diffusion Imaging-Based Investigation of Structural Connectivity
L. Bloy, M. Ingalhalikar, N. Batmanghelich
Brain Connectivity
[pdf]
Abstract
Structural connectivity models hold great promise for expanding what is known about the ways information travels throughout the brain. The physiologic interpretability of structural connectivity models depends heavily on how the connections between regions are quantified. This article presents an integrated structural connectivity framework designed around such an interpretation. The framework provides three measures to characterize the structural connectivity of a subject: (1) the structural connectivity matrix describing the proportion of connections between pairs of nodes, (2) the nodal connection distribution (nCD) characterizing the proportion of connections that terminate in each node, and (3) the connection density image, which presents the density of connections as they traverse through white matter (WM). Individually, each possesses different information concerning the structural connectivity of the individual and could potentially be useful for a variety of tasks, ranging from characterizing and localizing group differences to identifying novel parcellations of the cortex. The efficiency of the proposed framework allows the determination of large structural connectivity networks, consisting of many small nodal regions, providing a more detailed description of a subject’s connectivity. The nCD provides a gray matter contrast that can potentially aid in investigating local cytoarchitecture and connectivity. Similarly, the connection density images offer insight into the WM pathways, potentially identifying focal differences that affect a number of pathways. The reliability of these measures was established through a test/retest paradigm performed on nine subjects, while the utility of the method was evaluated through its applications to 20 diffusion datasets acquired from typically developing adolescents.
Generative-Discriminative Basis Learning for Medical Imaging
N. Batmanghelich, B. Taskar, C. Davatzikos
IEEE Trans Med Imaging
[pdf]
Abstract
This paper presents a novel dimensionality reduction method for classification in medical imaging. The goal is to transform very high-dimensional input (typically, millions of voxels) to a low-dimensional representation (small number of constructed features) that preserves discriminative signal and is clinically interpretable. We formulate the task as a constrained optimization problem that combines generative and discriminative objectives and show how to extend it to the semisupervised learning (SSL) setting. We propose a novel largescale algorithm to solve the resulting optimization problem. In the fully supervised case, we demonstrate accuracy rates that are better than or comparable to state-of-the-art algorithms on several datasets while producing a representation of the group difference that is consistent with prior clinical reports. Effectiveness of the proposed algorithm for SSL is evaluated with both benchmark and medical imaging datasets. In the benchmark datasets, the results are better than or comparable to the state-of-the-art methods for SSL. For evaluation of the SSL setting in medical datasets, we use images of subjects with Mild Cognitive Impairment (MCI), which is believed to be a precursor to Alzheimer’s disease (AD), as unlabeled data. AD subjects and Normal Control (NC) subjects are used as labeled data, and we try to predict conversion from MCI to AD on follow-up. The semi-supervised extension of this method not only improves the generalization accuracy for the labeled data (AD/NC) slightly but is also able to predict subjects which are likely to converge to AD.
2011
Regularized Tensor Factorization for Multi-Modality Medical Image Classification
N. Batmanghelich, B. Taskar, C. Davatzikos
International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2011)
[pdf]
Abstract
This paper presents a general discriminative dimensionality reduction framework for multi-modal image-based classification in medical imaging datasets. The major goal is to use all modalities simultaneously to transform very high dimensional image to a lower dimensional representation in a discriminative way. In addition to being discriminative, the proposed approach has the advantage of being clinically interpretable. We propose a framework based on regularized tensor decomposition. We show that different variants of tensor factorization imply various hypothesis about data. Inspired by the idea of multi-view dimensionality reduction in machine learning community, two different kinds of tensor decomposition and their implications are presented. We have validated our method on a multi-modal longitudinal brain imaging study. We compared this method with a publically available classification software based on SVM that has shown state-of-the-art classification rate in number of publications.
Disease Classification and Prediction via Semi-Supervised Dimensionality Reduction
N. Batmanghelich, D. Ye, K. Pohl, B. Taskar, C. Davatzikos
ISBI 2011 (Oral Presentation)
[pdf]
Abstract
We present a new semi-supervised algorithm for dimensionality reduction which exploits information of unlabeled data in order to improve the accuracy of image-based disease classification based on medical images. We perform dimensionality reduction by adopting the formalism of constrained matrix decomposition of [1] to semi-supervised learning. In addition, we add a new regularization term to the objective function to better capture the affinity between labeled and unlabeled data. We apply our method to a data set consisting of medical scans of subjects classified as Normal Control (CN) and Alzheimer (AD). The unlabeled data are scans of subjects diagnosedwith Mild Cognitive Impairment (MCI), which are at high risk to develop AD in the future. We measure the accuracy of our algorithm in classifying scans as AD and NC. In addition, we use the classifier to predict which subjects with MCI will converge to AD and compare those results to the diagnosis given at later follow-ups. The experiments highlight that unlabeled data greatly improves the accuracy of our classifier.
2010
Prediction of MCI Conversion via MRI, CSF Biomarkers, and Pattern Classification
C. Davatzikos, P. Bhatt, L. Shaw, N. Batmanghelich, J. Trojanowski
Neurobiology of Aging
[pdf]
Abstract
Magnetic resonance imaging (MRI) patterns were examined together with cerebrospinal fluid (CSF) biomarkers in serial scans of Alzheimer’s Disease Neuroimaging Initiative (ADNI) participants with mild cognitive impairment (MCI). The SPARE-AD score, summarizing brain atrophy patterns, was tested as a predictor of short-term conversion to Alzheimer’s disease (AD). MCI individuals that converted to AD (MCI-C) had mostly positive baseline SPARE-AD (Spatial Pattern of Abnormalities for Recognition of Early AD) and atrophy in temporal lobe gray matter (GM) and white matter (WM), posterior cingulate/precuneous, and insula. MCI individuals that converted to AD had mostly AD-like baseline CSF biomarkers. MCI nonconverters (MCI-NC) had mixed baseline SPARE-AD and CSF values, suggesting that some MCI-NC subjects may later convert. Those MCI-NC with most negative baseline SPARE-AD scores (normal brain structure) had significantly higher baseline Mini Mental State Examination (MMSE) scores (28.67) than others, and relatively low annual rate of Mini Mental State Examination decrease (-0.25). MCI-NC with midlevel baseline SPARE-AD displayed faster annual rates of SPARE-AD increase (indicating progressing atrophy). SPARE-AD and CSF combination improved prediction over individual values. In summary, both SPARE-AD and CSF biomarkers showed high baseline sensitivity, however, many MCI-NC had abnormal baseline SPARE-AD and CSF biomarkers. Longer follow-up will elucidate the specificity of baseline measurements.
Application of Trace-Norm and Low-Rank Matrix Decomposition for Computational Anatomy
N. Batmanghelich, A. Gooya, B. Taskar, C. Davatzikos
IEEE Computer Society Workshop on Mathematical Methods in Biomedical Image Analysis (MMBIA10)
[pdf]
Abstract
We propose a generative model to distinguish normal anatomical variations from abnormal deformations given a group of images with normal and abnormal subjects. We assume that abnormal subjects share common factors which characterize the abnormality. These factors are hard to discover due to large variance of normal anatomical differences. Assuming that the deformation fields are parametrized by their stationary velocity fields, these factors constitute a low-rank subspace (abnormal space) that is corrupted by high variance normal anatomical differences. We assume that these normal anatomical variations are not correlated. We form an optimization problem and propose an efficient iterative algorithm to recover the low-rank subspace. The algorithm iterates between image registration and the decomposition steps and hence can be seen as a group-wise registration algorithm. We apply our method on synthetic and real data and discover abnormality of the population that cannot be recovered by some of the well-known matrix decompositions (e.g. Singular Value Decomposition).